Developing a strategy to apply SNP molecular markers in winter oilseed rape DUS testing
Oilseed rape (Brassica napus) is the second most produced oilseed crop in the world and its cultivation is constantly increasing. Reflecting the growing economic importance of this species, the number of varieties listed in the French catalogue has increased significantly over the last ten years, mostly hybrids, resulting in a substantial increase in the size of the reference collection. To take into account the sensitivity of phenotypic characteristics due to environmental variations, the entire reference collection has to be redescribed every year during DUS testing. This represents huge technical and logistical challenges in terms of planning and infrastructure and significant costs. GEVES (France) and BSA (Germany) have therefore conducted a joint project with the objective of developing new approaches that combine genetic and phenotypic information to avoid having to redescribe the entire reference collection each year without compromising the quality of DUS tests. The new model should also be compatible with the different systems used in DUS to manage reference collections (GAIA or COY-D).
The objectives of the project were therefore :
– to genotype ~80% of the reference collection and produce a coherent molecular dataset for about 2000 winter oilseed rape varieties
– to use this molecular data to screen and optimise a set of 500 SNP markers
– to design and evaluate new approaches that combine genetic and phenotypic information based on SNP markers and historical field data accumulated over the last 10-15 years to optimise DUS trials.
This project allowed us to identify an optimised set of 360 SNPs with good genomic coverage and high discriminatory power. From this data, a new approach was developed that uses network analysis to define an ‘optimal reference collection’ (Figure 1). The proposed approach uses graph theory to detect groups of related varieties based on their proximity within a network describing the genetic relationships of the varieties to each other. In this model, algorithms are used to identify groups of varieties within the network based on their proximity (genetic similarity); only reference varieties in groups that also include candidate varieties will be included in the trials (Figure 1). Depending on the method used to manage reference collections (COY-D or GAIA), the model could reduce trial sizes by 20-45% in the first year of study, based on the preliminary genetic distance thresholds used to test the method. A schematic view of the implementation of the model is shown in Figure 2.
This optimisation method based on the network approach is implemented in R, an open source, cross-platform statistical tool. So far, the method has only been tested in silico, with genetic thresholds defined on the basis of the Franco-German collections. The molecular dataset currently represents ~80% of the Franco-German technical collections. A prerequisite for testing the model in situ will be to complete the genotyping of the reference collection and to evaluate the relevance of the proposed genetic thresholds. The evaluation of the model under real conditions should be the subject of a second project.
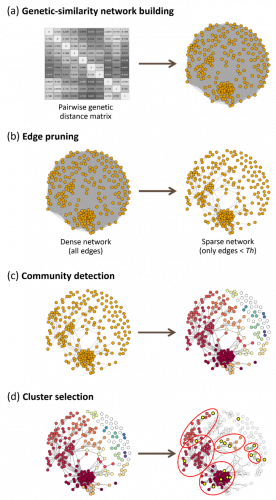
Figure 1: Description of the network approach. A matrix of genetic distances is calculated between all varieties and then transformed into a network, in which the nodes (varieties) are connected by links whose weight corresponds to the genetic distance between the nodes (a). All links with a genetic distance above a predefined threshold (GDTh) are removed to simplify the network (b) and a community detection algorithm is run to identify groups of related varieties (c). Only groups with at least one candidate variety (yellow circles) are included in the field trials (d).
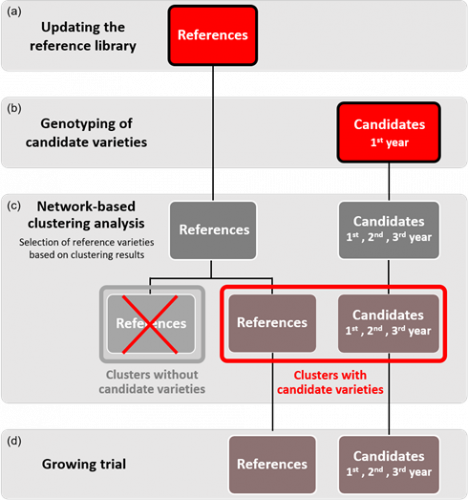
Figure 2: Schematic view of the proposed model for the integration of molecular data into the DUS test for winter oilseed rape varieties. Each year, the genotypes in the reference collection are updated to include newly added varieties (a). Once all applications have been received, the candidates are genotyped (b) and a genetic distance matrix is calculated between all varieties (candidates and varieties from the updated reference collection). The matrix is then analysed using the network approach which returns a table with the clustering results (c). Only groups with one or more candidate varieties are selected for inclusion in the growing trials (d).
Project co-financed by the CPVO and coordinated by GEVES in collaboration with Bundessortenamt.