A new RT-qPCR test for the identification of sugar beet yellows viruses
Sugar beet yellows is a complex of four viruses belonging to three viral genera that may be present in mono-infection or in co-infection:
- BYV (Beet Yellows Virus), belonging to the Closteroviruses

- BChV (Beet Chlorosis Virus) and BMYV (Beet Mild Yellowing Virus), belonging to the Polyeroviruses.
- BtMV (Beet Mosaic Virus), belonging to the Potyviruses.
This disease is mainly transmitted by the green peach aphid (Myzus percisae) and causes yield losses of up to 50%. Since 1993, aphid populations were controlled using neonicotinoids (NNI), however their use has been banned since 2018.
GEVES is coordinating a collaborative project of the National of Research and Innovation Plan (PNRI): Yellows Resisbeet, with two main objectives:
- To develop a protocol for evaluating sugar beet varietal resistance/tolerance to the four viruses responsible of yellows.
- To define one or more statistical models to estimate the productivity of varieties in trials in the presence of yellows.
Within the framework of this project, it is important to have an efficient detection test for the four viruses. Indeed, the ELISA method doesn’t allow to distinguish the two Polyeroviruses (BChV and BMYV). To answer this issue, the BioGEVES Detection Unit, in collaboration with INRAE of Colmar (Véronique Brault’s team – PNRI ProViBe project), has developed a detection and identification method for the four viruses responsible for beet yellows by multiplex RT-qPCR.
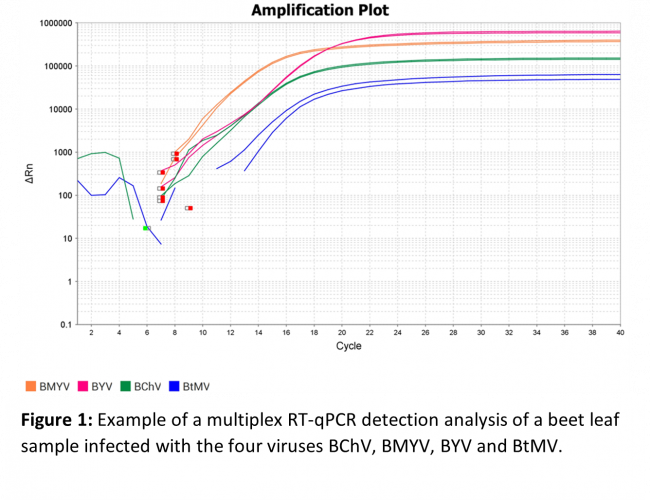
Four primer pairs and four probes TaqMan were designed to target respectively and specifically the four viruses BChV, BMYV, BYV and BtMV. The four TaqMan probes was each labeled with a different fluorophore, which allowed the development of a multiplex RT-qPCR method giving the ability to detect and identify all four viruses in a single RT-qPCR reaction. (Figure 1)
This method is in the final stage of validation, and performance criteria such as analytic specificity corresponding to the ability of a method to detect the target pest (inclusivity) while excluding non-target pests (exclusivity), and analytical sensitivity, corresponding to the smallest amount of target pest that can be detected, have been evaluated.
Thus, it was shown that the inclusivity and exclusivity rates are 100% for all primers and probes, except for the exclusivity rate of the BYV-targeting primers and probe which is 83%. The sensitivity of the multiplex RT-qPCR method was compared to the method currently used at GEVES, the ELISA, and it was demonstrated that multiplex RT-qPCR is 100 to 10,000 times more sensitive than ELISA depending on the virus.
Moreover, this multiplex RT-qPCR identification method was also shared with two Belgian research laboratories interested by this tool in the framework of their respective project:
- The Wallon Center for Agronomic Research (CRA-W), in the framework of the Virobeet project, which aims at studying the epidemics of different viruses of the beet and developing a management of beet yellows.
- Gembloux Agro Bio-Tech University of Liège, within the framework of a project also dealing with beet yellows in order to adapt the method to the detection and identification of the four viruses directly on aphids.
Once the method validation is finalized, it will be offered as a analytical service by the GEVES.
Publications in scientific revues are also envisaged in collaboration with our partners.